| | | |
LOCUS
|
ALLELE
|
PRED
|
|---|
Lab (country) | Period | Sample size |
p
l
| SD( p
l
) |
p
a
| SD( p
a
) |
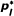
|
|---|
Lab 1 (Canada) | 97-02 | 116 (120) | 0.02373 | 0.00443 | 0.01422 | 0.00246 | 0.02825 |
Lab 2 (Iceland) | 03-05 | 60 | 0.00500 | 0.00288 | 0.00333 | 0.00166 | 0.00666 |
Lab 3 (Iceland) | 06 | 19 (20) | 0 | - | 0 | - | 0 |
Lab 4 (Norway) | 07-08 | 39 (40) | 0.00256 | 0.00256 | 0.00128 | 0.00128 | 0.00256 |
Total | 97-08 | 234 (240) | 0.01325 | 0.00236 | 0.00812 | 0.00131 | 0.01617 |
- The column "Period" shows the catch period for which each laboratory had responsibility for the NMDR. Except for the catches from 1997-98, which were genotyped in the year 2000, all individuals were genotyped the year following capture. The column "Sample size" contains the number of successfully regenotyped individuals from the laboratories, with the total number of regenotyping attempts in parenthesis. Initially 20 samples were selected from each year, but 6 samples had to be thrown out because of amplification failure. Locus EV001 was included for all probabilities of error in this table. In columns labeled LOCUS and ALLELE, p
l
and p
a
are the mean error rates (number of errors detected divided by number of loci/alleles), and SD(p) the corresponding empirical standard deviations. The column labeled PRED contains the predicted values,
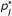 , of p
l
, calculated from p
a
using (2).
, of p
l
, calculated from p
a
using (2).
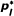
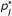 , of p
l
, calculated from p
a
using (2).
, of p
l
, calculated from p
a
using (2).